Abstract
Coronavirus disease 2019 (COVID-19) has rapidly become a global pandemic and no antiviral drug or vaccine is yet available for the treatment of this disease1,2,3. Several clinical studies are ongoing to evaluate the efficacy of repurposed drugs that have demonstrated antiviral efficacy in vitro. Among these candidates, hydroxychloroquine (HCQ) has been given to thousands of individuals infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)—the virus that causes COVID-19—worldwide but there is no definitive evidence that HCQ is effective for treating COVID-194,5,6,7. Here we evaluated the antiviral activity of HCQ both in vitro and in SARS-CoV-2-infected macaques. HCQ showed antiviral activity in African green monkey kidney cells (Vero E6) but not in a model of reconstituted human airway epithelium. In macaques, we tested different treatment strategies in comparison to a placebo treatment, before and after peak viral load, alone or in combination with azithromycin (AZTH). Neither HCQ nor the combination of HCQ and AZTH showed a significant effect on viral load in any of the analysed tissues. When the drug was used as a pre-exposure prophylaxis treatment, HCQ did not confer protection against infection with SARS-CoV-2. Our findings do not support the use of HCQ, either alone or in combination with AZTH, as an antiviral drug for the treatment of COVID-19 in humans.
Similar content being viewed by others
Main
Infection with SARS-CoV-2 is characterized by initial mild disease associated with respiratory symptoms at the peak of viral replication1,8. In some patients, a late severe immunological syndrome occurs 6–14 days after the onset of symptoms that may require intensive care and is responsible for most of the fatalities1,2,3.
HCQ has well-documented in vitro activity against various viruses4 and has emerged as an active compound against SARS-CoV-2 in different screening programmes, including a library of 1,520 Food and Drug Administration (FDA)-approved compounds5. In Vero E6 cells, HCQ has a 50% maximal effective concentration (EC50)5,9,10 that varies between 0.7 and 4 μM. It may inhibit viral transport in endosomes by alkalinizing the intra-organelle compartment10,11 and affect glycosylation, as reported for other viruses12. The drug may also act as an immunomodulatory agent13,14. In patients with lupus, HCQ decreases the level of inflammatory cytokines11,15,16, which may be relevant for the treatment of COVID-192. Furthermore, it has been proposed that AZTH, which displays in vitro antiviral activity against SARS-COV-25,17, could potentiate the efficacy of HCQ6. On the basis of these properties, HCQ has been considered for the treatment of COVID-19, alone or in combination with AZTH6,7.
We and others have set up non-human primate (NHP) models of SARS-CoV-2 infection18,19,20. Here we used cynomolgus macaques (Macaca fascicularis) to test different treatment strategies with HCQ, alone or in combination with AZTH, before or after the peak of viral replication. We also tested HCQ administration as pre-exposure prophylaxis treatment against SARS-CoV-2 infection.
In vitro efficacy of HCQ against SARS-CoV-2 infection
We first evaluated the in vitro antiviral activity of HCQ against a SARS-CoV-2 strain isolated from one of the first patients with COVID-19 in France. Post-infection treatment of Vero E6 cells with HCQ resulted in a dose-dependent antiviral effect, with 50% inhibitory concentration (IC50) values of 2.2 μM (0.7 μg ml−1) and 4.4 μM (1.4 μg ml−1) at 48 and 72 h after infection, respectively, which is within the range of previously reported values21 (Extended Data Fig. 1a). We next studied infection in a model of reconstituted human airway epithelium (MucilAir, Epithelix) developed from primary nasal or bronchial cells differentiated and cultured in an air–liquid interphase22. In contrast to previous observations for remdesivir23, the antiviral activity of HCQ in Vero E6 cells did not translate to the human airway epithelium model; doses of 1 μM or 10 μM HCQ did not significantly reduce SARS-CoV-2 apical viral titres at 48 h after infection (Extended Data Fig. 1b). HCQ also did not protect the integrity of epithelial tissue during infection, as the trans-epithelial electrical resistance values were comparable with the values of untreated cells and significantly lower than those of the mock-infected controls.
Infection of macaques with SARS-CoV-2
Cynomolgus macaques were infected on day 0 with a total dose of 106 plaque-forming units (PFU) of a primary SARS-CoV-2 isolate (BetaCoV/France/IDF/0372/2020; passaged twice in Vero E6 cells) by combined intranasal and intratracheal routes. Control NHPs (n = 8) had high viral loads in nasopharyngeal and tracheal samples (swabs), as estimated by quantitative PCR with reverse transcription (RT–qPCR), as early as 1 day after infection (d.p.i.). In tracheal samples, the viral load peaked at 2 d.p.i. (Fig. 1b and Extended Data Fig. 2a), with a median peak value of 7.9 log10 copies per ml. After 2 d.p.i., the viral loads progressively decreased and most NHPs had undetectable viral loads by 10 d.p.i. Similar profiles were observed for nasopharyngeal shedding (Extended Data Fig. 2b), whereas low viral loads were detected for more than 3 weeks in rectal samples and bronchoalveolar lavages (Extended Data Fig. 2c, d). NHPs exhibited mild clinical signs, including coughing or sneezing without dyspnoea, as has been reported for most patients with COVID-19 during the early infection period. The NHPs also developed early lymphocytopenia at 2 d.p.i. (Extended Data Fig. 5). No major changes were observed in heart rate, respiratory rate and oximetry analyses. Typical focal ground glass opacities associated with pleural thickening24,25 were observed in computed tomography (CT) scans with variable degrees of severity (Fig. 2 and Extended Data Fig. 3). Lesions were detectable as early as 2 d.p.i. and persisted up to 13 d.p.i. in some NHPs. None of the control NHPs developed a severe disease similar to what is observed in the late stages of the severe forms of the disease in humans.
a, Study design. The red dotted line indicates infection with 106 PFU of SARS-CoV-2 by the combined intranasal and intratracheal routes. Coloured areas indicate HCQ treatment periods. Each group received either a high (Hi) or a low (Lo) dose of HCQ according to the regimens described in the Methods. The treatment started 1 d.p.i. (D1) or 5 d.p.i. (D5), or 7 days before viral challenge for the pre-exposure prophylaxis (PrEP) group. One group received AZTH in combination with a high dose of HCQ. The control group received vehicle (water) as placebo. b–d, Viral loads were analysed by PCR in throat swabs (b, c) and bronchoalveolar lavages (BAL) (d). The limit of detection was estimated to be 2.3 log10 copies of SARS-CoV-2 RNA per ml and the limit of quantification was estimated to be 3.9 log10 copies per ml (dotted horizontal line). b, Shaded zones indicate treatment periods and each symbol and line combination represents one NHP. Dotted vertical lines indicate day of treatment initiation. c, d, Data are represented as medians of each group as described in a.
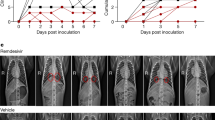
Lung lesions were assessed by chest CT before infection with SARS-CoV-2 and at 2, 5 and 11 or 13 d.p.i. a, Heat map of the overall CT score. Scores include lesion types (ground-glass opacity, crazy-paving pattern, consolidation or pleural thickening (scored from 0 to 3)) and lesion volume (scored from 0 to 4) summed for each lobe. Scores are consensus values from two independent evaluators. Dotted lines indicate treatment initiation. ‘X’ corresponds to missing data, macaques were not scanned at these time points. b, Representative images of lung lesions in two NHPs at baseline, 2, 5 and 13 d.p.i. Red arrows indicate typical lesions. Numbers at the bottom left of each image represent the CT score associated with the NHP and time point. Scores are the average over all scans made for the macaque at that time point.
Treatment with HCQ
To assess the anti-viral efficacy of HCQ, macaques received HCQ daily by gavage for 10 or more days. A treatment regimen of 90 mg kg−1 on 1 d.p.i. (loading dose) followed by a daily maintenance dose of 45 mg kg−1 was found to generate a clinically relevant plasma drug exposure in a group of uninfected NHPs (Extended Data Fig. 4b). In parallel, we also tested a lower treatment regimen, with a loading dose of 30 mg kg−1 and a maintenance dose of 15 mg kg−1. Overall, 9 NHPs were infected on day 0 and treated using the high treatment regimen (Hi D1, n = 5) or the low treatment regimen (Lo D1, n = 4), both starting at 1 d.p.i. We also examined the effect of a late low-dose treatment starting at 5 d.p.i.—when viral RNA levels are 3–4 log lower compared with peak values—to evaluate the benefit of HCQ in accelerating the clearance of the virus (Lo D5, n = 4). We focused on RT–qPCR analyses to assess the in vivo antiviral efficacy of HCQ because it provides a quantitative analysis, has a higher sensitivity and is less prone to variability than culture-based assays. In addition, RT–qPCR analysis is the only method that enables a comparison with results reported in human patients. Furthermore, virus titration in culture assays can be affected by many factors in addition to the number of viral particles, including any residual HCQ in the samples and host factors such as cytokines. All treated NHPs had tracheal viral RNA load kinetics that were similar to those of untreated NHPs, with median peak viral loads of 7.1 and 7.5 log10 copies per ml for the Hi D1 and Lo D1 groups, respectively, compared with 7.9 log10 copies per ml in the control group. Similarly, the areas under the curve (AUCs) of the viral load were similar between all groups, with values of 36.9 and 39.7 log10 copies × day per ml, for the Hi D1 and Lo D1 groups, respectively, compared with 40.3 log10 copies × day per ml in control NHPs (P = 0.62 and P = 0.37, respectively). Similar results were obtained for the nasopharyngeal swabs, and there were no differences in the levels of viral replication in bronchoalveolar lavages (Fig. 1d and Extended Data Fig. 2). In NHPs treated from 1 d.p.i. or 5 d.p.i., HCQ did not accelerate the time to viral clearance, and the median times to the first unquantifiable viral load were 4.5, 7.0, 7.0 and 7.0 days in the control, Lo D1, Hi D1 and Lo D5 groups, respectively.
Next, we evaluated the combination therapy of HCQ and AZTH, which was administered from 1 d.p.i., in which HCQ was given as a high dose as described above, and AZTH was given at a loading dose of 36 mg kg−1 followed by a daily dose of 18 mg kg−1 to mimic human exposure (Hi D1 + AZTH, n = 5). No effects of treatment were observed on either the viral RNA load in the different analysed tissues (Fig. 1d and Extended Data Fig. 2) or clinical scores. Clinical signs were comparable to control NHPs, with some NHPs exhibiting high CT scores in the Hi D1 + AZTH group (Fig. 2). In parallel, we also treated NHPs with a high dose of HCQ, starting 7 days before viral challenge as pre-exposure prophylaxis treatment (n = 5). Again, the kinetics of viral RNA loads were similar to those of the control group and no differences in the reduction in the AUC, peak viral load or time to first unquantifiable viral load were observed (Fig. 1 and Extended Data Fig. 2).
Relation between HCQ concentration and virus kinetics
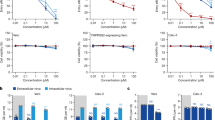
In the NHPs of Hi D1, Hi D1 + AZTH and pre-exposure prophylaxis groups, the plasma exposures were comparable to those observed in routine clinical practice 3–5 days after HCQ initiation using a dose of 200 mg three times daily (Fig. 3a). Drug trough concentrations were lower in both the Lo D1 and Lo D5 groups. When we assessed whether the higher drug exposure could generate more-rapid virus clearance, neither the time to attain the viral load limit of quantification nor the peak viral load were significantly associated with plasma HCQ concentrations (Fig. 3b–d). Finally, in an additional group of uninfected macaques, we characterized the HCQ pharmacokinetics in blood and plasma as the accumulation of HCQ in the lungs 6 days after the initiation of treatment (Fig. 3e, f and Extended Data Fig. 4). The blood concentrations in the high-dose HCQ group were higher than 1.4 μg ml−1, showing that the drug concentrations in the blood remained above the drug EC50 values that we identified in Vero E6 cells during in vitro efficacy assessment of HCQ against SARS-CoV-2 infection. The mean blood-to-plasma ratio was 6.8 (Extended Data Fig. 4), close to the value of 7.2, which was reported in healthy volunteers during various treatment intervals and durations26. Consistent with predictions made in physiological pharmacokinetic models, these levels of drug exposure in the plasma and blood produced higher exposure concentrations in lung tissues, with a lung-to-plasma ratio ranging from 27 to 177 (Fig. 3f), allowing lung tissues to achieve concentrations that were mostly above the drug EC50 values found in Vero E6 cells in all NHPs during the treatment period.
a, Individual mean plasma trough concentrations of HCQ in NHPs during treatment. HCQ plasma trough concentrations determined within the context of routine therapeutic drug monitoring using the same method are provided for hospitalized patients (n = 25) who received 200 mg three times daily. The box bounds represent the 25th and 75th percentiles, the middle line shows the mean, and whiskers show the minimum and maximum values. b, Time to the first measurement below the limit of quantification in NHPs with a mean plasma trough concentration <0.1 μg ml−1 (black) or >0.1 μg ml−1 (grey). The time to viral suppression was compared between the two groups using a log-rank test (n = 31 macaques in total). c, Peak viral load according to mean HCQ plasma trough concentration. d, Area under the curve (AUC) or viral kinetic curve between 1 and 9 d.p.i. c, d, A Spearman correlation test was performed to assess the association between drug concentration and viral kinetic parameters (n = 31 macaques in total). e, HCQ lung and plasma concentrations in uninfected NHPs (n = 6). f, HCQ lung and blood concentrations in uninfected NHPs (n = 6).
Pathogenesis and host response to HCQ treatment
High alanine aminotransferase and creatinine kinase levels were observed in NHPs treated with the high HCQ and particularly the HCQ + AZTH regimen compared with control NHPs (Extended Data Figs. 7, 8).
HCQ treatment did not prevent lymphocytopenia (Extended Data Fig. 5) nor pulmonary lesions, as shown by CT scan analysis (Fig. 2 and Extended Data Fig. 3). Similar lesion scores were observed in control and treated NHPs.
All NHPs exhibited an increase in the concentrations of type-I IFNα, IL-1RA, CCL2 and CCL11 in plasma at 2 d.p.i. (Fig. 4 and Extended Data Fig. 6). In addition, IL-15 peaked early during infection, which suggests that innate lymphoid cells have a role in the control of initial viral replication in both drug-treated and untreated NHPs. When compared with control NHPs, TNF was significantly increased and IL-1RA was significantly reduced at 2 d.p.i. (Fig. 4 and Extended Data Fig. 6) in the groups that received the high dose of HCQ alone (P = 0.032 and P = 0.028, respectively) or with AZTH (P = 0.037 and P = 0.045, respectively).
a, Heat map of plasma concentrations of eotaxin (also known as CCL-11), MCP-1 (also known as CCL-2), IFNα, IL-1RA, IL-2 and IL-15 at 0, 2, 5, 7 and 9 d.p.i. The asterisk indicates a significant difference in IL-1RA concentrations at 2 d.p.i. between the control group and the Hi D1 and Hi D1 + AZTH groups (P = 0.0287 and P = 0.0451, respectively). Further analyses of IL-1RA are shown in Extended Data Fig. 6c. Statistical significance was determined using a two-sided Mann–Whitney U-test without correction for multiple testing.
Conclusions
Our study shows that cynomolgus macaques are a relevant model for the analysis of the early stages of SARS-Cov-2 infection in humans1,3,18,19,20,27,28. We found no antiviral activity nor clinical efficacy of HCQ treatment, regardless of the timing of treatment initiation, either before infection, early after infection (before the peak of the viral load) or late after infection (after the peak of the viral load). This was in spite of high HCQ concentrations in the blood and lungs, and plasma exposures that were similar to those observed in patients with COVID-19 treated who were with HCQ. Thus, treatment with HCQ is unlikely to have antiviral activity in respiratory compartments. Our results illustrate the frequent discrepancy between results from in vitro assays and in vivo experiments, as reported for other viral infections such as influenza, dengue or chikungunya virus, for which clinical trials did not demonstrate efficacy of chloroquine or HCQ for the treatment of these infections4,29.
In conclusion, our evaluation of HCQ in the NHP model does not support its use as an antiviral agent for the treatment of COVID-19 in humans.
Methods
Ethics and biosafety statement
Cynomolgus macaques (M. fascicularis), aged 37–40 months and originating from Mauritian AAALAC-certified breeding centres, were used in this study. All macaques were housed in IDMIT infrastructure facilities (CEA, Fontenay-aux-Roses), under BSL-2 and BSL-3 containment when necessary (animal facility authorization D92-032-02, Prefecture des Hauts de Seine, France) and in compliance with European Directive 2010/63/EU, the French regulations and the Standards for Human Care and Use of Laboratory Animals of the Office for Laboratory Animal Welfare (OLAW, assurance number A5826-01, United States). The protocols were approved by the institutional ethical committee ‘Comité d’Ethique en Expérimentation Animale du Commissariat à l’Energie Atomique et aux Energies Alternatives’ (CEtEA 44) under statement number A20-011. The study was authorized by the ‘Research, Innovation and Education Ministry’ under registration number APAFIS#24434-2020030216532863v1.
HCQ and AZTH
Hydroxychloroquine sulfate (HCQ) was manufactured for Sanofi by the Chinoin Pharmaceutical and Chemical Works under good manufacturing practice conditions and provided as the base powder. Batch number DU017 was solubilized extemporaneously in water at 5, 10 or 15 mg ml−1 depending on the group and the dose. Tablets of AZTH (250 mg) (Sandoz, batch number KH5525) were crushed and suspended extemporaneously at 12 mg ml−1 of AZTH in water.
Macaques and study design
To evaluate the efficacy of HCQ and HCQ + AZTH treatments, the macaques were randomly assigned in sex-balanced experimental groups. No statistical methods were used to predetermine sample size. Challenged macaques were exposed to a total dose of 106 PFU of SARS-CoV-2 through a combination of intranasal and intratracheal routes (day 0), using atropine (0.04 mg kg−1) as premedication and ketamine (5 mg kg−1) with medetomidine (0.042 mg kg−1) as anaesthesia. The regimen comprising a high dose of HCQ in group ‘Hi D1’ (n = 5) consisted of a loading dose of 90 mg kg−1 at 1 d.p.i. and a daily maintenance dose of 45 mg kg−1, for a total of 10 days. The ‘Hi D1 + AZTH’ regimen (n = 5) consisted of the same HCQ regimen as for the Hi D1 group combined with one loading dose of 36 mg kg−1 of AZTH at 1 d.p.i., followed by a daily maintenance dose of 18 mg kg−1 AZTH for 10 days. The low-dose (Lo) regimen consisted of a HCQ loading dose of 30 mg kg−1 and a daily maintenance dose of 15 mg kg−1 for 12 days. The low-dose treatment of the ‘Lo D1’ group (n = 4) was initiated at 1 d.p.i. and the low-dose treatment of the ‘Lo D5’ group (n = 4) was initiated at 5 d.p.i. The PrEP regimen (n = 5) consisted of a loading dose of 30 mg kg−1 HCQ 7 days before challenge, followed by a daily dose of 15 mg kg−1 for 4 days and 45 mg kg−1 for 3 days before virus challenge and then 45 mg kg−1 until 6 d.p.i. Treatments were delivered by gavage. Placebo-treated macaques received water, which was the vehicle for HCQ. Macaques were observed daily and clinical examinations were performed at baseline, daily for one week and then twice weekly on macaques that were anaesthetized using ketamine (5 mg kg−1) and metedomidine (0.042 mg kg−1). Body weight, rectal temperature, respiration, heart rates and oxygen saturation were recorded and blood, as well as nasopharyngeal, tracheal and rectal swabs, were collected. Bronchoalveolar lavages were performed using 50 ml sterile saline on 6, 14, 21 and 28 d.p.i. Chest CT scans were performed at baseline and on 2, 5 and 11 or 13 d.p.i. in macaques that were anaesthetized using tiletamine (4 mg kg−1) and zolazepam (4 mg kg−1). Blood cell counts, haemoglobin and haematocrit were determined from EDTA-treated blood samples using a HMX A/L analyser (Beckman Coulter). Biochemistry parameters including alanine aminotransferase (ALT), aspartate aminotransferase (AST), albumin, haptoglobin, creatinine, creatine kinase, lactate dehydrogenase (LDH) and total protein, were analysed with standard kits (Siemens) and C-reactive protein with a canine kit (Randox) in lithium heparin plasma, inactivated with Triton X-100, using an ADVIA1800 analyser (Siemens).
The pharmacokinetics of HCQ was assessed using the same administration procedure in six uninfected macaques, randomly assigned as pairs into three experimental groups as described in Extended Data Fig. 4. The pharmacokinetic low (PK Lo) group received a low loading dose (30 mg kg−1) at day 0 and a low daily maintenance dose (15 mg kg−1) for 5 days. The pharmacokinetic high (PK Hi) and ‘PK Hi + AZTH’ groups received a high loading HCQ dose (90 mg kg−1) on day 0 and a high daily maintenance dose (45 mg kg−1) for 6 days, without or with AZTH (loading dose of 36 mg kg−1 and maintenance of 18 mg kg−1), respectively. Blood samples were taken at 0, 2, 4 and 6 h after treatment on day 0, and before treatment on the following days. For the PK Hi and PK Hi + AZTH groups, blood samples were also collected at 0, 2, 4 and 6 h after treatment after treatment on day 5. Macaques were humanly euthanized 24 h after the administration of the last dose using 18.2 mg kg−1 of pentobarbital sodium intravenously under tiletamine (4 mg kg−1) and zolazepam (4 mg kg−1) anaesthesia. Samples of lung were collected at necropsy for HCQ quantification.
Determination of HCQ concentrations
Quantification of HCQ in plasma, blood and lung tissues was performed by a sensitive and selective validated high-performance liquid chromatography coupled with tandem mass spectrometry method (Quattro Premier XE LC-MS/MS, Waters) as previously described30, with lower limits of quantification of 0.015 μg ml−1 for plasma and 0.05 μg ml−1 for blood and lung tissue. Blood samples were centrifuged within 1 h to collect plasma samples. Lung biopsies collected after euthanasia were thoroughly rinsed with cold 0.9% NaCl to remove blood contamination and blotted with filter paper. Then, each lung biopsy was weighed and homogenized with 1 ml of 0.9% NaCl using a Mixer mill MM200 (Retsch). Cellular debris was removed by centrifugation, and the supernatant was stored at −80 °C.
HCQ was extracted by a simple protein precipitation method, using methanol for plasma and ice-cold acetonitrile for blood and tissue homogenates. In brief, 100 μl of sample matrix was spiked with 10 μl of internal standard working solution (HCQ-d5, Alsachim), vortexed for 2 min followed by centrifugation for 10 min at 4 °C. The supernatant was evaporated for blood- and tissue-homogenate samples. Dry residues or plasma supernatants were then transferred to 96-well plates and 5 μl was injected. To assess the selectivity and specificity of the method and matrix effect, blank plasma, blood and tissues from control macaques were processed and compared with that of HCQ and index-selectivity-spiked plasma, blood or tissue homogenate samples. Furthermore, each baseline sample (H0) of treated macaques was processed in duplicate, including one spiked with HCQ prepared equivalent to quality control samples.
Concentrations in blood (μg ml−1), plasma (μg ml−1) and lung (μg g−1) were determined for each uninfected macaque and in plasma only for infected macaques. Drug accumulation in the lung was assessed by calculating a lung-to-blood and a lung-to-plasma concentration ratio. No signs of haemolysis were observed, either visually (when only plasma samples were available) or after verification of the consistency between the two matrixes (when both plasma and blood samples were available).
HCQ plasma trough concentrations determined within the context of routine therapeutic drug monitoring using the same method, 3–5 days after initiation of HCQ at 200 mg three times daily were provided for comparison.
Viruses and cells
For the in vivo studies, SARS-CoV-2 virus (hCoV-19/France/lDF0372/2020 strain) was isolated by the National Reference Center for Respiratory Viruses (Institut Pasteur) as previously described31. Virus stocks used in vivo were produced by two passages on mycoplasma-free Vero E6 cells in Dulbecco’s modified Eagle’s medium (DMEM) without FBS, supplemented with 1% penicillin (10,000 U ml−1) and streptomycin (10,000 μg ml−1) and 1 μg ml−1 TPCK-trypsin at 37 °C in a humidified CO2 incubator and titrated on Vero E6 cells.
For the in vitro studies, the viral strain hCoV-19/France/IDF0571/2020 was provided by X. Lescure and Y. Yazdanpanah from the Bichat Hospital, where the isolate was obtained from a patient returning from Jichang (China) and passaged three times. For the virus used in the in vivo experiments, whole-genome sequencing was performed as previously described31 with no modifications observed compared with the initial specimen27. For sequencing of the virus used in vitro, viral RNA extraction was done using the QiAmp viral RNA Kit (Qiagen). The complete viral genome sequence was obtained using Illumina MiSeq sequencing technology. Sequences were deposited after assembly on the GISAID EpiCoV platform under accession numbers EPI_ISL_406596 for hCoV-19/France/lDF0372/2020 and EPI_ISL_411218 for hCoV-19/France/IDF0571/2020.
Viral replication kinetics and antiviral treatment in Vero E6 cells
Vero E6 cells were seeded 24 h in advance in multi-well 6 plates, washed twice with PBS and then infected with SARS-CoV-2 at the indicated multiplicities of infection (MOI). For HCQ treatment, the inoculum of infected Vero E6 cells was removed 1 h after infection (h.p.i.) and cells were immediately treated with solutions in DMEM of HCQ. Supernatants were collected at 48 and 72 h.p.i. and stored at −80 °C for RNA extraction and viral quantification.
Viral quantification in Vero E6 cells
Viral stocks and collected samples were titrated by tissue-culture infectious dose 50% (TCID50 ml−1) in Vero E6 cells, using the Reed and Muench statistical method. Relative quantification of the viral genome was performed by one-step real-time quantitative reverse transcriptase and polymerase chain reaction (RT–qPCR) from viral RNA extracted using the QiAmp viral RNA Kit (Qiagen) in the case of supernatants or apical washings. Primer and probe sequences were selected from those designed by the School of Public Health/University of Hong Kong (L. Poon, D. Chu and M. Peiris) and synthetized by Eurogentec23. Real-time one-step RT–qPCR was performed using the EXPRESS One-Step Superscript qRT–PCR Kit (Invitrogen, 1178101K). Thermal cycling was performed in a StepOnePlus Real-Time PCR System (Applied Biosystems) in MicroAmp Fast Optical 96-well reaction plates (Applied Biosystems, 4346907), as previously described23.
Viral infection and treatment in reconstituted human airway epithelia
MucilAir human airway epithelia (HAE) reconstituted from human primary cells obtained from nasal (pool of donors) or bronchial (single donors) biopsies were provided by Epithelix and maintained in air–liquid interphase with specific culture medium in Costar Transwell inserts (Corning) according to the manufacturer’s instructions. For infection experiments, apical poles were gently washed twice with warm OptiMEM medium (Gibco, ThermoFisher Scientific) and then infected directly with a 150-μl dilution of virus in OptiMEM medium, at a MOI of 0.1. For mock infection, the same procedure was performed using OptiMEM as inoculum. Samples collected from apical washes or basolateral medium at different time points were separated into two tubes: one for TCID50 viral titration and one RT–qPCR. HAE cells were collected in RLT buffer (Qiagen) and total RNA was extracted using the RNeasy Mini Kit (Qiagen) for subsequent RT–qPCR and Nanostring assays. Treatments with HCQ were applied through basolateral poles. All treatments were initiated on day 0 (1 h after viral infection) and continued once daily. Samples were collected at 48 h.p.i. Variations in trans-epithelial electrical resistance (ΔTEER) were measured using a dedicated volt–ohm meter (EVOM2, Epithelial Volt/Ohm Meter for TEER) and expressed as Ω cm−2.
Virus quantification in NHP samples
Upper respiratory (nasopharyngeal and tracheal) and rectal specimens were collected with swabs (Universal transport medium, Copan; or Viral Transport Medium, CDC, DSR-052-01). Tracheal swabs were performed by insertion of the swab above the tip of the epiglottis into the upper trachea at approximately 1.5 cm of the epiglottis. All specimens were stored between 2 °C and 8 °C until analysis with a plasmid standard concentration range containing an rdrp gene fragment including the RdRp-IP4 RT–PCR target sequence. The protocol describing the procedure for the detection of SARS-CoV-2 is available on the WHO website (https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf?sfvrsn=3662fcb6_2).
Plasma cytokine analysis
Cytokines were quantified in EDTA-treated plasma using NHP ProcartaPlex immunoassay (ThermoFisher Scientific) for IFNα, IL-1RA, IL-1β, CCL-2 (also known as MCP-1), CCL-11 (also known as eotaxin), CXCL-11 (also known as ITAC), CXCL-1 (also known as BLC), granzyme B and PDGF-BB, using NHP Milliplex (Millipore) for CD40L, G-CSF, GM-CSF, IFNγ, IL-2, IL-4, IL-5, IL-6, CXCL-8 (also known as IL-8), IL-10, IL-13, IL-15, IL-17A, CCL-3 (also known as MIP-1α), CCL-4 (also known as MIP-1β), TNF, VEGF and a Bioplex 200 analyser (Bio-Rad) according to manufacturer’s instructions.
Chest CT and image analysis
Acquisition was done using a CT system (Vereos-Ingenuity, Philips) in BSL-3 containment facilities on anaesthetized macaques placed in a supine position and monitored for heart rate, oxygen saturation and body temperature. An intravenous bolus of iodine contrast agent (Vizipaque, 320mg ml−1, GE Heathcare, 3 ml kg−1) was injected (Medrad CT Stellant injector, Bayer) in the saphenous vein 20 s before the initiation of CT scan acquisition. The CT detector collimation was 64 × 0.6 mm, the tube voltage was 120 kV and intensity of about 120 mA. Automatic dose optimization tools (Dose Right, Z-DOM, 3D-DOM; Philips Healthcare) regulated the intensity. CT images were reconstructed with a slice thickness of 1.25 mm and an interval of 0.25 mm.
Images were analysed using INTELLISPACE PORTAL 8 software (Philips healthcare). All images had the same window level of −300 and window width of 1,600. Lesions were defined as ground glass opacitiy, crazy-paving pattern, consolidation or pleural thickening as previously described24,25. Lesions and scoring were assessed independently in each lung lobe by two individuals, and the final results were made by consensus. The overall CT score includes lesion type (scored from 0 to 3) and lesion volume (scored from 0 to 4) summed for each lobe as described in Extended Data Fig. 3.
Statistical analysis
The following viral kinetic parameters were calculated in each experimental group as medians (and minimum–maximum): viral load peak, area under the curve of the log10 viral load, time to first unquantifiable viral load. Each viral kinetic parameter was compared with untreated macaques using Wilcoxon rank-sum or log-rank tests (Microsoft Excel 2016, GraphPad Prism version 7). To evaluate a potential effect of drug exposure on viral dynamics, we further evaluated the correlation of the viral kinetic parameters with the plasma concentrations of HCQ, taking the mean trough concentrations observed in each infected macaque between 1 and up to 15 days after treatment as a marker of drug exposure during treatment period (Spearman test, without adjusting for multiple testing).
Ethics committee
All information on the ethics committee is available at https://cache.media.enseignementsup-recherche.gouv.fr/file/utilisation_des_animaux_fins_scientifiques/22/1/comiteethiqueea17_juin2013_257221.pdf.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
The data that support the findings of this study are included in the paper and Supplementary Information.
References
He, X. et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 26, 672–675 (2020).
Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020).
Chen, G. et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 130, 2620–2629 (2020).
Touret, F. & de Lamballerie, X. Of chloroquine and COVID-19. Antiviral Res. 177, 104762 (2020).
Touret, F. et al. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 10, 13093 (2020).
Gautret, P. et al. Clinical and microbiological effect of a combination of hydroxychloroquine and azithromycin in 80 COVID-19 patients with at least a six-day follow up: a pilot observational study. Travel Med. Infect. Dis. 34, 101663 (2020).
Magagnoli, J. et al. Outcomes of hydroxychloroquine usage in United States veterans hospitalized with Covid-19. Med 1, 1–14 (2020).
Li, Q. et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 382, 1199–1207 (2020).
Wang, M. et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30, 269–271 (2020).
Liu, J. et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 6, 16 (2020).
Ponticelli, C. & Moroni, G. Hydroxychloroquine in systemic lupus erythematosus (SLE). Expert Opin. Drug Saf. 16, 411–419 (2017).
Savarino, A., Di Trani, L., Donatelli, I., Cauda, R. & Cassone, A. New insights into the antiviral effects of chloroquine. Lancet Infect. Dis. 6, 67–69 (2006).
Rainsford, K. D., Parke, A. L., Clifford-Rashotte, M. & Kean, W. F. Therapy and pharmacological properties of hydroxychloroquine and chloroquine in treatment of systemic lupus erythematosus, rheumatoid arthritis and related diseases. Inflammopharmacology 23, 231–269 (2015).
Schrezenmeier, E. & Dörner, T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat. Rev. Rheumatol. 16, 155–166 (2020).
Monzavi, S. M. et al. Efficacy analysis of hydroxychloroquine therapy in systemic lupus erythematosus: a study on disease activity and immunological biomarkers. Inflammopharmacology 26, 1175–1182 (2018).
Fanouriakis, A. et al. 2019 Update of the Joint European League Against Rheumatism and European Renal Association-European Dialysis and Transplant Association (EULAR/ERA-EDTA) recommendations for the management of lupus nephritis. Ann. Rheum. Dis. 79, 713–723 (2020).
Zhang, B. et al. Macrolide derivatives reduce proinflammatory macrophage activation and macrophage-mediated neurotoxicity. CNS Neurosci. Ther. 25, 591–600 (2019).
Rockx, B. et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 368, 1012–1015 (2020).
Williamson, B. N. et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature https://doi.org/10.1038/s41586-020-2423-5 (2020).
Shi, R. et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124 (2020).
Yao, X. et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 71, 732–739 (2020).
Nicolas de Lamballerie, C. et al. Characterization of cellular transcriptomic signatures induced by different respiratory viruses in human reconstituted airway epithelia. Sci. Rep. 9, 11493 (2019).
Pizzorno, A. et al. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell. Rep. Med 1, 100059 (2020).
Pan, F. et al. Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19). Radiology 295, 715–721 (2020).
Shi, H. et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 20, 425–434 (2020).
Tett, S. E., Cutler, D. J., Day, R. O. & Brown, K. F. A dose-ranging study of the pharmacokinetics of hydroxy-chloroquine following intravenous administration to healthy volunteers. Br. J. Clin. Pharmacol. 26, 303–313 (1988).
Liu, Y. et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 20, 656–657 (2020).
Chen, N. et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395, 507–513 (2020).
Roques, P. et al. Paradoxical effect of chloroquine treatment in enhancing chikungunya virus infection. Viruses 10, 268 (2018).
Chhonker, Y. S., Sleightholm, R. L., Li, J., Oupický, D. & Murry, D. J. Simultaneous quantitation of hydroxychloroquine and its metabolites in mouse blood and tissues using LC-ESI-MS/MS: an application for pharmacokinetic studies. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1072, 320–327 (2018).
Lescure, F. X. et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 20, 697–706 (2020).
Acknowledgements
We thank B. Delache, E. Burban, J. Demilly, N. Dhooge, S. Langlois, P. Le Calvez, M. Potier, F. Relouzat, J. M. Robert and C. Dodan for help with animal studies; B. Fert and C. Mayet for help with in vivo imaging studies; Q. Pascal for performing the necropsies; J. Morin for help with the cytokine measurements and preparation of reagents; K. Barthelemy, M. Basso, N. Doudka and M. Giocanti for help with HCQ concentration measurements; B. Lacarelle and R. Guilhaumou for help with analysing internal drug concentration data; J. Bertrand for help with analysing pharmacokinetic data; D. Desjardins for help with the AZTH pharmacokinetic study; C. Aubenque, M. Barendji, L. Bossevot, N. Dimant, J. Dinh, A. S. Gallouet, M. Leonec, I. Mangeot and K. Storck for help with sample processing; M. Albert, M. Barbet and F. Donati for help with the production, titration and sequencing of the virus stocks used in vivo and processing of samples for RT–PCR; A. S. Gallouet, S. Keyser, E. Marcos-Lopez, B. Targat and B. Vaslin for help with the experimental studies in the context of COVID-19-induced constraints; F. Ducancel and Y. Gorin for help with the logistics and safety management; E. Sultan for providing guidance on HCQ dose selection and discussion on pharmacokinetics/pharmacodynamics results and commenting on the paper; Sanofi for providing the HCQ batch used in these experiments; the Fondation Bettencourt Schueller and the Region Ile-de-France for the contribution to the implementation of imaging facilities; and the Domaine d’Intérêt Majeur (DIM) ‘One Health’ for its support. This study received financial support from REACTing, the National Research Agency (ANR; AM-CoV-Path) and the European Union’s Horizon 2020 (H2020) research and innovation program Fight-nCov (101003555), European Union IMI2 program CARE (101005077) and the European Infrastructure TRANSVAC2 (730964). The virus stock was obtained through the EVAg platform (https://www.european-virus-archive.com/), funded by H2020 (653316). The Infectious Disease Models and Innovative Therapies (IDMIT) research infrastructure is supported by the ‘Programme Investissements d’Avenir’, managed by the ANR under reference ANR-11-INBS-0008.
Author information
Authors and Affiliations
Contributions
A.B. performed RT–PCR viral quantification and analysed the data. A.G. contributed to statistical analysis. A.P. performed in vitro evaluation of HCQ (Vero E6 and HAE), and contributed to data analysis and the preparation of the manuscript. B.H. contributed to study design, data analysis and the writing of the paper. B.L. coordinated the in vitro evaluation of HCQ (Vero E6 and HAE), analysed the data and contributed to the writing of the paper. C.C. coordinated the imaging facility. J.G. contributed to data analysis, the pharmacokinetics/pharmacodynamics study and the writing of the paper. J.L. contributed to clinical follow-up of macaques, data analysis and the writing of the paper. M.R.C. designed the in vitro evaluation of HCQ (Vero E6 and HAE), supervised and coordinated the work, analysed the data and contributed to the writing of the paper. N.D.-B. contributed to the animal work and cytokine measurements, analysed the data and coordinated IDMIT core activities. N.E. developed the RT–qPCR assay and analysed the data. N.K. performed CT scans and acquisition parameter design, and contributed to data analysis. O.T. performed in vitro evaluation of HCQ (Vero E6 and HAE), contributed to data analysis and manuscript preparation. P.M. contributed to project conception and design of the study, contributed to animal work, the coordination of the experiments, data analysis and the writing of the paper. R.H.T.F. coordinated the animal core facility, and contributed to study design and data analysis. R.L.G. conceived the project, designed the study, coordinated the work, analysed the data and wrote the article. R.M. contributed to the design of the study, animal work, data analysis and contributed the writing of the paper. S.B. performed RT–qPCR viral quantification and analysed the data. C.S. supervised and coordinated the HCQ pharmacokinetics analysis, provided clinical data on plasma HCQ levels and contributed to the writing of the paper. F.T. contributed to in vitro antiviral evaluation. S.v.d.W. conceived the project, designed the study, provided the viral challenge stock, coordinated the viral load quantification, analysed the data and wrote the paper. T.N. performed CT scans and quantification, contributed to the quantification design, generated CT figures and wrote the paper. V.C. contributed to data analysis, statistical analyses, figures design and the writing of the paper. V.E. developed the RT–qPCR viral quantification assay. X.d.L. contributed to study design, pharmacokinetics/pharmacodynamics analysis and the writing of the paper.
Corresponding author
Ethics declarations
Competing interests
J.G. has worked as consultant for Roche.
Additional information
Peer review information Nature thanks Robin Ferner and Debby van Riel for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 In vitro evaluation of the antiviral activity of HCQ against SARS-CoV-2.
a, Dose–response curves of HCQ at 48 and 72 h.p.i. in Vero E6 cells. Vero E6 cells were seeded 24 h in advance in multi-well six-well plates, washed twice with PBS and then infected with SARS-CoV-2 (BetaCoV/France/IDF0571/2020 SARS-CoV-2 strain) at the indicated MOI. The inoculum of infected Vero E6 cells was removed 1 h.p.i. and cells were immediately treated with different concentrations of HCQ. Supernatants were collected at 48 and 72 h.p.i. and stored at −80 °C for RNA extraction and viral titration by RT–qPCR. Results were expressed in relative viral production compared with the untreated control. The table summarizes the IC50, cytotoxic concentration 50% (CC50) and selectivity index (SI) for each condition. b, Apical relative viral production and trans-epithelial resistance (TEER in Ohms cm−2) between the apical and basal poles in nasal and bronchial HAE at 48 h.p.i. MucilAir HAE reconstituted from human primary cells obtained from nasal or bronchial biopsies were provided by Epithelix and maintained in air–liquid interphase. For infection experiments, apical poles were gently washed twice with warm OptiMEM medium (Gibco, ThermoFisher Scientific) and then infected directly with nasal swab samples or a 150-μl dilution of virus in OptiMEM medium, at a MOI of 0.1. For mock infection, the same procedure was performed using OptiMEM as inoculum. Samples collected from apical washes were separated into two tubes: one for TCID50 viral titration and one for RT–qPCR. Results are expressed in relative viral production compared with the infected untreated control. The table summarizes the relative viral production values (mean ± s.d.) and the infectious titres (log10[TCID50 ml−1) of three biological replicates tested in duplicate.
Extended Data Fig. 2 Viral loads of SARS-CoV-2-infected cynomolgus macaques treated with HCQ.
a–d, Viral loads measured by RT–qPCR in throat swabs (a), nasal swabs (b), rectal swabs (c) and bronchoalveolar lavages (d). The limit of detection was estimated at 2.3 log10 copies of SARS-CoV-2 RNA per ml and the limit of quantification was estimated at 3.9 log10 copies per ml (dotted horizontal line). Shaded zones indicate treatment periods. Baseline was adjusted to day 0 on the graphs. CTRL, control; D1, treatment started on day 1; D5, treatment started on day 5; Hi, high HCQ dose; Lo, low HCQ dose; PrEP, pre-exposure prophylaxis.
Extended Data Fig. 3 Representative transversal slices of lung CT scans from SARS-CoV-2-infected cynomolgus macaques treated with HCQ.
Imaging was performed at baseline and day 2, 5 and 11 or 13 post-exposure to SARS-CoV-2. a–f, Images are presented for each macaque according to their group, with a window level of −300 and a window width of 1,600. g, h, CT scoring scales (g) and example of the analysis worksheet (h). Total CT score is the sum of all lung lobe scores including lesion type, extension contributions and pleural effusion severity. ND, not determined.
Extended Data Fig. 4 Plasma and blood HCQ concentrations of six uninfected NHPs.
a, Pharmacokinetic (PK) study design. Three groups of two cynomolgus macaques received either a high (Hi) dose regimen of HCQ composed of a loading dose of 90 mg kg−1 and a daily maintenance dose of 45 mg kg−1 or a low (Lo) dose regimen composed of a 30 mg kg−1 loading dose and a daily maintenance dose of 15 mg kg−1. One group received AZTH in combination with HCQ with a loading dose of 36 mg kg−1 followed by an 18 mg kg−1 daily maintenance dose. b, Individual plasma and blood HCQ concentrations (conc.) up to 5–6 days after the initiation of treatment. c, Correlation between plasma and blood HCQ concentrations.
Extended Data Fig. 5 Complete blood count of SARS-CoV-2-infected cynomolgus macaques treated with HCQ.
a–d, Absolute numbers of white blood cells (WBC) (a), lymphocytes (b), neutrophils (c) and monocytes (d) in SARS-CoV-2-infected macaques. Baseline was adjusted to day 0 on the graphs, and to the first day of treatment (day 7 pre-exposure) for the PrEP group. Shaded zones indicate treatment periods.
Extended Data Fig. 6 Cytokines and chemokines in the plasma of SARS-CoV-2-exposed cynomolgus macaques treated with HCQ.
a, b, Heat maps of the plasmatic concentrations of 20 cytokines. Each column represents one cytokine or chemokine; the colour scale (in pg ml−1) is shown at the bottom. The asterisk indicates a significant difference in the concentration of TNF at 2 d.p.i. between the control group and the Hi D1 and/or Hi D1 + AZTH groups, as shown in d. ‘X’ indicates that no measurement was determined for this time point. c, d, Concentrations of IL-1RA and TNF at 2 d.p.i. Each plot represents one macaque. Symbols represent mean of duplicate measurements for individual macaques of two replicates for IL-1RA and a single replicate for TNF. Statistical significance was determined using a two-sided Mann–Whitney U-test without correction and P values are indicated on the graphs (c, d).
Extended Data Fig. 7 Plasma ALT levels of cynomolgus macaques treated with HCQ.
In addition to the six SARS-CoV-2-infected groups, three groups of two macaques were treated but not infected to follow HCQ pharmacokinetics. a–f, ALT levels in the plasma are shown for all infected macaques (a–e) and the uninfected, HCQ-treated macaques (f). Shaded zones indicate treatment periods.
Extended Data Fig. 8 Biochemistry analysis of cynomolgus macaques treated with HCQ.
In addition to the six SARS-CoV-2-infected groups, three groups of two macaques were treated but not infected to follow HCQ pharmacokinetics. a–f, AST, albumin, creatine kinase, creatinine, haptoglobin, LDH, C-reactive protein (CRP) and total proteins levels were analysed in the plasma of all infected (a–e) and uninfected, treated (f) groups. Shaded zones indicate treatment periods.
Supplementary information
Rights and permissions
About this article
Cite this article
Maisonnasse, P., Guedj, J., Contreras, V. et al. Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature 585, 584–587 (2020). https://doi.org/10.1038/s41586-020-2558-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2558-4
This article is cited by
-
Whole-body visualization of SARS-CoV-2 biodistribution in vivo by immunoPET imaging in non-human primates
Nature Communications (2025)
-
Fetal Zika virus inoculation in macaques revealed control of the fetal viral load during pregnancy
Virology Journal (2024)
-
Multi-omics analysis uncovered systemic lupus erythematosus and COVID-19 crosstalk
Molecular Medicine (2024)
-
Hydroxychloroquine and COVID-19 story: is the low-dose treatment the missing link? A comprehensive review and meta-analysis
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)
-
Immunogenicity and efficacy of VLA2001 vaccine against SARS-CoV-2 infection in male cynomolgus macaques
Communications Medicine (2024)